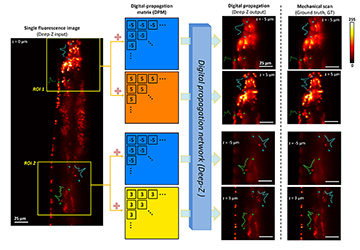
 Deep-Z 3D digital refocusing of a C. elegans fluorescence image provides results comparable to those obtained by more labor- and photon-dose-intensive mechanical scanning, thus reducing imaging time and potential photodamage to the sample. [Enlarge figure]
Deep-Z 3D digital refocusing of a C. elegans fluorescence image provides results comparable to those obtained by more labor- and photon-dose-intensive mechanical scanning, thus reducing imaging time and potential photodamage to the sample. [Enlarge figure]
High-throughput volumetric fluorescence microscopic imaging remains an important challenge in biology, life sciences and engineering. Imaging 3D fluorescent samples usually involves scanning to acquire images at different focal planes, which unavoidably reduces the imaging speed and increases photon dose on the sample due to repeated exposure and excitation. While a number of methods have achieved scanning-free volumetric fluorescence microscopic imaging,1–3 these earlier techniques typically require adding customized optical components, which cause photon loss and increased aberrations.
As an alternative approach, we have created a deep-learning-based framework, Deep-Z, to digitally refocus 2D fluorescence microscopic images onto user-defined 3D surfaces.4 This framework enables volumetric inference from a standard wide-field fluorescence microscope image without any mechanical scanning, additional hardware or trade-offs between imaging resolution and speed. In Deep-Z, a convolutional neural network (CNN) is trained to take as its input both a snapshot 2D fluorescence image and a user-defined digital propagation matrix (DPM) that represents per-pixel axial refocusing distances. The CNN outputs a digitally refocused image at the exact 3D surface defined by the DPM.4
Using Deep-Z, we imaged the neurons of C. elegans nematodes in 3D using a standard wide-field fluorescence microscope, and computationally extended its native depth-of-field by 20-fold. At the output of Deep-Z, non-distinguishable fluorescent features in the input image can be brought into focus and resolved at different depths according to their true axial positions in 3D, matching focus-scanned ground truth without performing actual mechanical scanning. This significantly reduces the imaging time and photodamage to the sample.
Deep-Z further enabled high-throughput longitudinal volumetric reconstruction of
C. elegans neuronal activities at up to 100 Hz. A temporally synchronized virtual image stack of a moving C. elegans was generated from a sequence of 2D images, captured by a standard wide-field fluorescence microscope at a single focal plane.4 Spatially non-uniform DPMs can also be input into CNN to digitally correct for sample drift, tilt, field-curvature and other image aberrations, after the acquisition of a single image.4
Deep-Z is further capable of performing virtual refocusing across different microscopy modalities.4 For instance, the framework can be trained to virtually refocus a snapshot wide-field fluorescence image, and blindly infer and output a confocal-equivalent image at a different focus. This is especially useful for reducing the imaging time and photodamage that might occur during confocal scanning.
Researchers
Yichen Wu, Yair Rivenson, Hongda Wang, Yilin Luo, Eyal Ben-David, Laurent A. Bentolila, Christian Pritz and Aydogan Ozcan, University of California, Los Angeles, CA, USA
References
1. R. Prevedel et al. Nat. Methods 11, 727 (2014).
2. S. Abrahamsson et al. Opt. Express 23, 7734 (2015).
3. J. Rosen et al. Nat. Photon. 2, 190 (2008).
4. Y. Wu et al. Nat. Methods 16, 1323 (2019).
