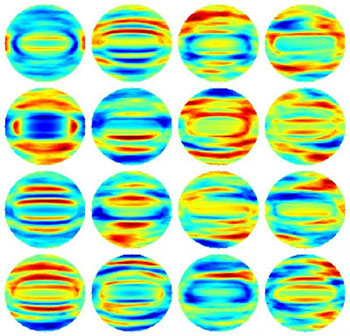
Fourier transform light scattering (FTLS) maps of bacterial samples from 16 experiments. Maps such as these were used to create a “classification model” that would allow a computer algorithm to recognize specific bacterial species, in near real time, based on their light-scattering behavior. [Image: Young-Ju Jo and YongKeun (Paul) Park, Korea Advanced Institute of Science and Technology]
In a novel mash-up of standard microscopy, advanced photonics, and machine learning, a team of South Korean scientists has devised a new approach for identifying dangerous bacteria that is potentially cheaper and far more rapid than conventional lab culturing techniques (Opt. Express, doi: 10.1364/OE.23.015792). The scientists believe that the system could form the basis for new point-of-care diagnostics, especially in resource-poor settings, and also could find use in screening food for contamination.
At present, the “gold standard” for bacterial identification, involving culture of the bacteria from blood samples, requires days to produce a measurable signal. Even the fastest techniques, involving real-time quantitative polymerase chain reaction and gene sequencing, can take hours. That timescale is too slow for some diseases, such as sepsis, which can kill in a matter of hours—and which, as a result, are commonly treated using the inefficient and potentially dangerous “nuclear option” of broad-spectrum antibiotics. While optical techniques for bacterial identification have previously been floated to address the speed issue, most require an additional step of labeling with chemical agents such as nanoparticles, an awkward requirement in the clinic.
To get to a fast and label-free optical approach, scientists led by YongKeun Park of the Korea Advanced Institute of Science and Technology (KAIST) adopted a two-step approach: First, adapt standard microscopes to take a high-fidelity optical “fingerprint” of the suspect bacteria; and, second, use machine-learning algorithms—the same kind deployed in face-recognition software—to tie that fingerprint to the known optical signature of a specific bacterial species in a library of samples.
For the first step, the researchers adapted a standard light microscope to perform quantitative phase imaging (QPI), using a 523-nm laser as the illumination source and tying in additional optical elements for the interferometry. They recorded holograms of isolated known bacteria, and extracted amplitude and phase images of the bacteria from the holograms. The team then applied Fourier transform light scattering (FTLS) numerical modeling to develop 2-D light-scattering maps of the bacterial species based on the measured light fields. The 2-D maps provide a proxy for the behavior of light scattering within the bacteria, according to the scientists, because they’re based on 3-D variations of refractive index that are tied to internal structure and composition.
For the second, machine-learning step, the team used a raft of experimentally generated FTLS light-field maps from specific, previously identified bacteria to “train” a computer to recognize bacteria based on the optical signature. The result was a classification model against which real-world samples could be measured. The scientists tested the system by creating such a model for four rod-shaped bacterial species that overlap considerably in size and shape, and that are thus very difficult to distinguish based on surface morphology alone. The system was able to correctly suss out the optical fingerprint of the bacteria, and thus correctly identify the bacterial species, with an overall accuracy rate of 94 percent.
Team leader YongKeun Park believes that the system, which involves a low-cost modification to a standard microscope and a “single-shot” measurement, could prove extremely useful in clinical and food-safety work, especially in rural areas and the developing world. Indeed, in a separate paper late in 2014 (Opt. Lett., doi: 10.1364/OL.39.003630), Park and KAIST colleague KyeoReh Lee unveiled a compact QPI unit that can “convert a simple existing microscope into a holographic one,” and that thus could be used to capture light-field images in a wide variety of settings for use in fingering bacterial culprits. The team plans to field-test the new technique in Tanzania in the coming weeks.

