
Digital holography is in many ways similar to classic holography, except that optoelectronic sensors are used instead of photographic film. This allows for numerical processing of holographic data for a host of applications in biomedicine, public health and environmental monitoring. This technique has been adapted to microscopy, and it is widely used in the three-dimensional (3-D) visualization of biological specimens.
The essence of holographic imaging lies in the fact that, when coherent light propagates through a semi-transparent object, its amplitude and phase get modulated due to light-matter interaction. As a result, the outgoing wavefront carries information about the entire 3-D structure of the object.
With digital holographic microscopy (DHM), one can indirectly record information about the phase and amplitude of the object’s wavefront and thus numerically reconstruct sectional images of biological specimens at different depths from only a single holographic sampling. DHM is therefore commonly categorized as a form of 3-D optical-computational microscopy.
At its core, DHM includes an optical interferometer, which is used to form an interference pattern between the Fresnel diffraction of cells or microorganisms and a reference wavefront. The intensity distribution of this interference pattern is digitized using an optoelectronic image sensor array (e.g., CCD or CMOS camera), and the result is a digital hologram in which the information about the 3-D structure of a biological specimen is encoded. An inverse Fresnel transform can be used to decode the digital hologram and to reconstruct the specimen’s characteristic phase and amplitude modulation information. DHM does not require mechanical scanning and thus offers an advantage over laser scanning and confocal 3-D microscopy methods, which commonly require beam steering in the transverse plane and/or mechanical translation of the specimen along the optical axis in order to bring different planes of the specimen into focus.
Conventional bright-field optical microscopes typically map optical absorption across the specimen and dismiss the phase information. Due to the low absorption coefficient of cytoplasm (the jelly-like substance that fills the cell) and most cellular components, the resulting 2-D bright-field images of cells are often low-contrast and produce few useful details.
One of the typical practices used to increase the contrast of semitransparent cells is to stain and fix them with high absorption coefficient dyes. Unfortunately, this is an invasive process that often terminates the cell’s life cycle. However, cell biologists typically need to monitor live cells to study their behavior and dynamics. Fortunately, light phase is very sensitive to the refractive index mismatch between the cytoplasmic content and the surrounding medium. This property has been leveraged in conventional phase contrast as well as differential interference contrast (DIC) microscopic techniques to create high-contrast intensity images.
However, such techniques do not provide optical thickness information about cells. Although phase contrast and DIC techniques have been very useful in the past, the qualitative data that are generated by such techniques does not lend itself to quantitative characterization and the automatic recognition of cells and microorganisms. In addition, in phase contrast and DIC modalities, one needs to mechanically scan the specimen in order to bring different sections into focus. This adds to the imaging time and forbids real-time cell inspection and dynamic events over time.
DHM is a quantitative phase imaging method that directly and noninvasively provides the optical thickness of cells in live, dynamic conditions. Optical thickness is related to cell thickness and its 3-D shape through the cell refractive index.
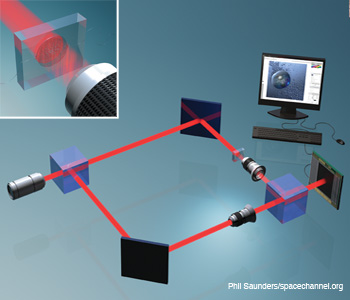 Digital holographic microscopy (DHM) integrated with a microfluidic device for 3-D imaging, identification and dynamic analysis of cells and microorganisms. For Gabor mode DHM, the reference arm of the interferometer is removed.
Digital holographic microscopy (DHM) integrated with a microfluidic device for 3-D imaging, identification and dynamic analysis of cells and microorganisms. For Gabor mode DHM, the reference arm of the interferometer is removed.
Thus, when combined with appropriate computational algorithms, DHM is an attractive candidate for non-invasive real-time 3-D imaging and for identifying cells and microorganisms. A digital hologram contains rich, quantitative information about the 3-D structure of the specimen, which can be captured in one exposure and used to identify cells and recognize objects. To this end, one can leverage the wealth of pattern recognition techniques that have been developed for automatic object recognition and used in applications in medicine, military, robotics and various industries.
Recently, researchers have proposed using 3-D sensing and imaging systems for noninvasive automated identification of cells based on their 3-D structure and dynamics. Real-time, automated screening, analysis and identification of biological specimens can aid in disease diagnostics, environmental monitoring and early detection of pandemics. By contrast, conventional biological characterization methods are typically invasive, labor-intensive and time-consuming. They also require staining, which can be very detrimental to the cells. Moreover, they might not be effective in large-scale deployment. In addition, specimen analysis based on 2-D features such as the shape, size and morphology of a biological specimen is not always conclusive.
Therefore, the integration of 3-D computational imaging, information optics and DHM bears promise for a reliable, automated and low-cost tool for rapid sensing, visualization and identification of cells (for example, blood or stem cells), which can be used to detect and track disease states.
Single-exposure DHM
In single-exposure DHM, one records a digital Fresnel hologram from the specimen in either on-axis or off-axis configurations. In on-axis mode, the real and conjugate images are superimposed on the digital hologram. Separating them requires a spatial or temporal phase-shifting method, which adds to the complexity and capture time. By introducing a small angular separation between the two interfering beams in off-axis mode, one effectively separates the real and conjugate images in a single exposure at the expense of suboptimal use of sensor space-bandwidth product. This separation allows one to discard the virtual image and perform quantitative phase measurements based on the real image (or vice versa).
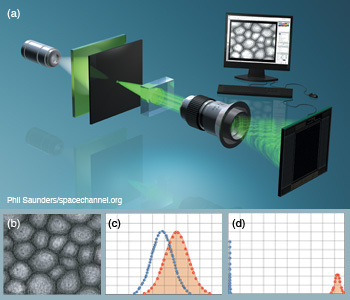 Gabor digital holographic microscopy. (a) DHM with Gabor configuration under partially coherent illumination for cell identification. (b) Digital hologram of a sunflower stem cell reconstructed after applying Gabor wavelet filtering. (c, d) Statistical sampling distributions show improved separation between nontraining true and false class samples when (d) Gabor-filtered digital holograms are used instead of (c) non-filtered holograms. Null hypothesis is the training true class.
Gabor digital holographic microscopy. (a) DHM with Gabor configuration under partially coherent illumination for cell identification. (b) Digital hologram of a sunflower stem cell reconstructed after applying Gabor wavelet filtering. (c, d) Statistical sampling distributions show improved separation between nontraining true and false class samples when (d) Gabor-filtered digital holograms are used instead of (c) non-filtered holograms. Null hypothesis is the training true class.
However, single-exposure, on-axis DHM is simpler to implement, and it allows for the use of partially coherent light instead of a fully coherent source. In the partially coherent case, there is no need for a separate reference beam, since the semitransparent specimen transmits part of the incident wave unperturbed. In other words, the reference and object arms of the interferometer can be collapsed into one. This mode is referred to as Gabor holography.
In addition, the microscope objective that is used for magnifying the field can be removed in a lensless implementation. The divergence of the incident beam allows for some geometrical magnification, and the numerical processing of the hologram replaces the image forming role of the microscope objective. Although not applicable to quantitative phase measurement, single-exposure, on-axis Gabor holographic microscopy has been shown to work reasonably well for 3-D cell identification, with a large gain in simplicity, compactness and expense that is very valuable for point-of-care health solutions.
With either on-axis or off-axis configurations, the digital holograms are recorded by a CCD sensor and transferred to a computer for processing. One can then reconstruct the specimen’s 3-D image, or a stack of 2-D images, from the recorded digital hologram by inverse Fresnel transformation. For the automated identification of biological specimens, signal processing and/or statistical pattern recognition algorithms can be used to extract the unique features of the cell under inspection from its reconstructed complex-valued images.
For instance, a segmentation algorithm may be used to separate the cells from the background in the reconstructed image. After 3-D segmentation, sampling feature vectors can be generated by randomly extracting the complex pixel values in the segmented 3-D image. Alternatively, one can directly apply pattern recognition analysis to the digital hologram data to identify the cells. This approach would remove the 3-D holographic reconstruction step, making the computations faster.
A number of statistical classification algorithms are developed particularly for the identification of biological specimens. Cross-comparison with 2-D intensity images has shown that complex 3-D images that are reconstructed from digital holograms provide more discriminating features and allow for better classification of cells.
One way to simplify the system is to implement a Gabor-mode DHM with a partially coherent source. In this configuration, ballistic photons automatically provide the reference beam. The photons pass through the specimen and its surrounding medium without any scattering. The use of partially coherent sources such as LEDs (as opposed to laser beams) provides a lower cost and more compact 3-D imaging instrument.
To improve identification and recognition, one may apply wavelet transformation to the recorded digital hologram prior to reconstruction in order to extract the discriminating features of the cells under inspection. Unlike the Fourier transform family, which solely provides frequency analysis, wavelets provide space-frequency analysis that is beneficial in the study of complex objects involving local features.
Another interesting application of DHM is in the analysis of embryonic stem cells, which are being explored for their potential as a therapeutic solution in regenerative medicine. It is important for biologists to conduct quantitative monitoring in order to better understand stem cells’ characteristics over their development cycle in the colony. Combined with statistical analysis tools, DHM is a useful inspection modality for stem cell applications due to its noninvasiveness and recognition capabilities. This combination can be used for monitoring the progression of stem cell colonies and quantifying their evolution and differentiation over time.
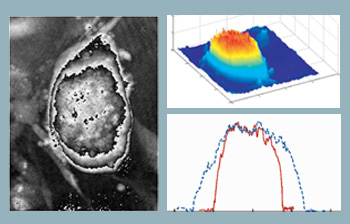 Optical thickness of embryonic stem cells. Computed phase distribution of a three-day-old embryonic stem cell colony using DHM. Microscope objective is 20X with 0.4 NA. (Left) Computed phase distribution and (center) 3-D optical path length distribution. (Right) Cell thickness profile along x (solid line) and y (dotted line) directions.
Optical thickness of embryonic stem cells. Computed phase distribution of a three-day-old embryonic stem cell colony using DHM. Microscope objective is 20X with 0.4 NA. (Left) Computed phase distribution and (center) 3-D optical path length distribution. (Right) Cell thickness profile along x (solid line) and y (dotted line) directions.
 Integrated optofluidic DHM system. (Left) Example of digital hologram of Euglena Acus, (center) intensity and (right) unwrapped phase reconstruction of the wavefront.
Integrated optofluidic DHM system. (Left) Example of digital hologram of Euglena Acus, (center) intensity and (right) unwrapped phase reconstruction of the wavefront.
The figure to the right shows the computed phase distribution of a three-day-old stem cell colony. Time-lapsed holographic imaging can provide information on the growth of the colony in both lateral and axial directions. One application in this area is to use the tool for real-time and automated classification of differentiated stem cells and the fibroblast feeder layer cells at the cell harvesting stage.
DHM with optofluidic devices
The advent of microfluidic devices has opened a new frontier in biological research, owing to the precise control and high throughput that these devices offer. An integrated system of miniaturized micro-channels, culture sites and nutrient reservoirs can be designed and fabricated to manipulate small volumes of biological specimens—often at the individual cell level. Many of these advantages can be carried over to a DHM-based system with the goal of screening, classifying and manipulating cells in a mixed culture.
The microfluidic device can be designed as a complex network of micro-channels with multiple inlet and outlet ports controlled by precision pumps. The micro-channels fabricated this way can be as small as 20 mm in width and 100 mm in depth—a size that would allow cells to pass one by one for inspection.
The light passing through the device is modulated by both the cells and inhomogeneities of the silicon polymer used to fabricate the microfluidic device. In addition, the micro-channel exhibits a lensing effect that distorts the wavefront further. However, digital holographic interferometry allows one to digitally isolate the device wavefront modulation from that of the cells. As a result, one can accurately reconstruct the optical path length distribution of an organism based on wavefront phase reconstruction.
 Single beam three dimenisonal microscopy. Single beam recording for complex wavefront reconstruction and quantitative phase contrast three-dimensional microscopy.
Single beam three dimenisonal microscopy. Single beam recording for complex wavefront reconstruction and quantitative phase contrast three-dimensional microscopy.
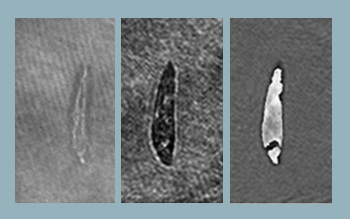
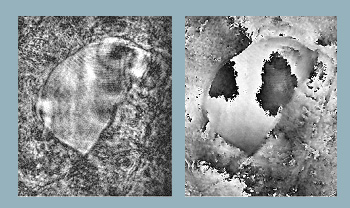 Reconstructed object wavefront using single beam. Object is a section of an insect wing. (Left) Reconstructed intensity and (right) reconstructed phase.
Reconstructed object wavefront using single beam. Object is a section of an insect wing. (Left) Reconstructed intensity and (right) reconstructed phase.
The 3-D reconstruction of cells can be used with statistical pattern recognition algorithms to recognize and classify the cells in real-time. The precision control feature of microfluidic devices can then be leveraged to act on the cell classification results in order to physically segregate different types of cells in a mixed culture at a two-way split channel.
Toward single beam quantitative 3-D imaging
DHM in Gabor mode does not provide quantitative phase information because the real and conjugate wavefronts, as well as the undiffracted reference fields, are all spatially overlapped in the hologram. One can use a two-beam, off-axis interferometer to separate the three field components. However, this approach results in inefficient use of the sensor’s space-bandwidth product and can introduce challenges for some applications, including the use of sources with exotic wavelengths or low coherence, and in miniaturized instrumentation due to the difficulty involved in matching the two optical path distances.
These problems can be remedied by reconstructing a complex wavefront with a single beam; this is done by using a volume speckle field, making multiple recordings and applying phase-retrieval algorithms. The complex wavefront can be reconstructed by taking multiple intensity samplings, after the object wavefront has been converted into a volume speckle field with rapid but detectable intensity variations and then applying scalar diffraction integral.
By using multiple constraints, one can obtain fast convergence and accurate wavefront reconstruction. The use of volume speckle field makes the implementation compact, and the method is attractive for low coherence applications. A single-beam technique can be implemented with as few as three sampling planes in a small package—an advantage over the bulkier off-axis DHM used for quantitative phase measurement.
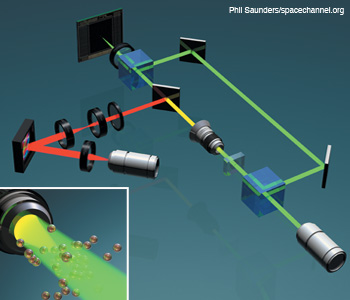 Integration of DHM and optical tweezers in a Mach–Zehnder configuration. DHM and HOT can be integrated for imaging and dynamic optical cell trapping using a phase-only spatial light modulator. The dichroic mirror reflects at the trapping beam wavelength.
Integration of DHM and optical tweezers in a Mach–Zehnder configuration. DHM and HOT can be integrated for imaging and dynamic optical cell trapping using a phase-only spatial light modulator. The dichroic mirror reflects at the trapping beam wavelength.
Integration with optical tweezers
Another fertile area is the integration of optical tweezers technology with DHM. In fact, the two technologies are synergistic with respect to screening and classification tasks. They are also easy to integrate in one compact system. Holographic optical tweezers (HOT), in particular, provide a versatile and flexible tool for rapid micromanipulation of cells through active control of a phase-only spatial light modulator. Unlike conventional optical tweezing, the holographic technique allows one to simultaneously generate and manipulate multiple traps in 3-D space and time.
The figure to the right illustrates how the two modalities can be combined in one optical system. In the more practical single-beam configuration, HOT requires a high NA objective, which in turn reduces the depth of field for bright field microscopy. However, it is challenging to achieve successful trapping without proper imaging. In the integrated system, the DHM can overcome this problem through its inherent ability to digitally reconstruct the wavefront within an extended depth of field and track the specimen in 3-D to provide feedback for active trapping.
Researchers have shown that normal and cancerous cells can be classified, noninvasively, based on the trapping force exerted on them by the optical trap. This force can be calculated based on cell escape-velocity measurements. This additional parameter provided by a dynamic HOT system can augment statistical pattern-recognition-based cell identification using DHM to create a more robust classifier.
Looking ahead
We are taking the path towards a more quantitative approach to challenges in biological research. 3-D optical-computational microscopy can provide a number of advantages over traditional imaging modalities in terms of visualization, quantitative inspection, recognition and automatic classification of unicellular species.
Every day brings more challenging requirements in biological processing; we need new tools that offer high throughput, automatic and quantitative inspection, and manipulability. Three-dimensional computational imaging such as DHM integrated with appropriate numerical algorithms is a promising candidate. We hope this article will generate discussions and encourage the optics and computational imaging communities to take part in cross-disciplinary collaborations in this exciting field.
I. Moon wishes to acknowledge support from the Mid-career Researcher Program through the National Research Foundation of Korea funded by the Ministry of Education, Science and Technology (2010-0010391). B. Javidi wishes to acknowledge support under the Humboldt Foundation. Arun Anand wishes to acknowledge UGC and DST, Govt. of India for project grants.
Inkyu Moon is with the School of Computer Engineering, Chosun University, in Gwangju, South Korea. Mehdi Daneshpanah is with GE Global Research, One Research Circle, Niskayuna, N.Y., U.S.A. Arun Anand is with the applied physics department, faculty of technology and engineering, The MS University of Baroda, Vadodara, India. Bahram Javidi is with the department of electrical and computer engineering at the University of Connecticut, Storrs, Conn., U.S.A.
References and Resourcess
>> D. Gabor. "A new microscopic principle," Nature 161, 777-8 (1948).
>> E.N. Leith and J. Upatnieks. "Reconstructed Wavefronts and Communication Theory," J. Opt. Soc. Am. 52, 1123-30 (1962).
>> J.W. Goodman and R. W. Lawrence. "Digital image formation from electronically detected holograms," Appl. Phys. Lett. 11, 77-9 (1967).
>> I. Yamaguchi and T. Zhang. "Phase-shifting digital holography," Opt. Lett. 22, 1268-70 (1997).
>> D. Frank Dubois et al. "Improved Three-Dimensional Imaging with a Digital Holography Microscope with a Source of Partial Spatial Coherence," Appl. Opt. 38, 7085-94 (1999).
>> E. Cuche et al. "Digital holography for quantitative phase-contrast imaging," Opt. Lett. 24, 291-3 (1999).
>> B. Javidi and E. Tajahuerce. "Three dimensional object recognition using digital holography," Opt. Lett. 25, 610-12 (2000).
>> D.B. Murphy, Fundamentals of light microscopy and electronic imaging, Wiley-Liss, N.Y., U.S.A., 2001.
>> W. Xu et al. "Digital in-line holography for biological applications," Proc. Natl. Acad. Sci. 98, 11301-5 (2001).
>> G. Pedrini and H.J. Tiziani. "Short-coherence digital microscopy by use of a lensless holographic imaging system," Appl. Opt. 41, 4489 (2002).
>> J. Jang and B. Javidi. "Three-dimensional integral imaging of micro-objects," Opt. Lett. 29, 1230-2 (2004).
>> L. Repetto et al. "Lensless digital holographic microscope with light-emitting diode illumination," Opt. Lett. 29, 1132-4 (2004).
>> Y. Zhang et al. "Reconstruction of in-line digital holograms from two intensity measurements," Opt. Lett. 29, 1787-9 (2004).
>> B. Javidi et al. "Three-dimensional imaging and recognition of microorganism using single-exposure on-line (SEOL) digital holography," Opt. Express 13, 4492-6 (2005).
>> P. Ferraro et al. "Extended focused image in microscopy by digital holography," Opt. Express 13, 6738-49 (2005).
>> T. Kreis, ed., Handbook of Holographic Interferometry, Wiley, N.Y., U.S.A., 2005.
>> P. Marquet et al. "Digital holographic microscopy: a noninvasive contrast imaging technique allowing quantitative visualization of living cells with subwavelength axial accuracy," Opt. Lett. 30, 468-70 (2005).
>> B. Javidi et al. "Three-dimensional identification of biological microorganism using integral imaging," Opt. Express 14, 12095-107 (2006).
>> Y. Frauel et al. "Three Dimensional Imaging and Display Using Computational Holographic Imaging," Proc. IEEE 94, 636-54 (2006).
>> I. Moon and B. Javidi. "3-D identification of stem cells by computational holographic imaging," J.R. Soc. Interface 4, 305-13 (2007).
>> S.-H. Lee and D.G. Grier. "Holographic microscopy of holographically trapped three-dimensional structures," Opt. Express 15, 1505-12 (2007).
>> E.A. Ozcan and U. Dmirci. "Ultrawide-field lens-free monitoring of cells on-chip," Lab. Chip. 8, 98-106 (2008).
>> I. Moon and B. Javidi. "3-D visualization and identification of biological microorganisms using partially temporal incoherent light in-line computational holographic imaging," IEEE Trans. on Medical Imaging 27, 1782-90 (2008).
>> F. Schaal et al. "Marker-free cell discrimination by holographic optical tweezers," J. Eur. Opt. Soc.- Rap. Public. 4, 09028 (2009).
>> I. Moon et al. "Automated three-dimensional identification and tracking of micro/nanobiological organisms by computational holographic microscopy," Proc. IEEE 97, 990-1010, (2009).
>> M. Levoy et al. "Recording and controlling the 4d light field in a microscope using microlens arrays," J. Microsc. 235, 144-62 (2009).
>> A. Anand and B. Javidi. "Three dimensional microscopy with single beam wavefront sensing and reconstruction from speckle fields," Opt. Lett. 35, 766-8 (2010).
>> M. Daneshpanah et al. "3D holographic imaging and trapping for noninvasive cell identification and tracking," IEEE J. Display Technol. 6, 490-9 (2010).
>> D. Shin et al. "Optofluidic system for three-dimensional sensing and identification of micro-organisms with digital holographic microscopy," Opt. Lett. 35, 4066-8 (2010).
